
是一家鸵鸟相关产业开发与销售为一体的企业
 热线电话:
热线电话:

 热线电话:
热线电话:
NEWS CENTER
了解公司较新资讯
Whole Genome Characterization and Genetic Evolution Analysis of a New Ostrich Parvovirus Kunpeng Yuan 1 , Dongdong Wang 2 , Qingdong Luan 1 , Ju Sun 2 , Qianwen Gao 1 , Zhiyao Jiang 1, Shouchun Wang 1 , Yijun Han 1 , Xueting Qu 1 , Yueying Cui 1 , Shimei Qiu 1 , Youxia Di 3, Xiaoyi Wang 3 , Shige Song 3 , Peiheng Wang 3 , Shilong Xia 3 , Yongle Yu 4 , Weiquan Liu 4,* and Yanbo Yin 1,* 1 College of Veterinary Medicine, Qingdao Agricultural University, Qingdao 266019, China; 18306390700@163.com (K.Y.); lqddmj@126.com (Q.L.); gqwdmj2020@163.com (Q.G.); jzydmj@sina.com (Z.J.); wangshouchun2011@sina.com (S.W.); hyjdmj@souhu.com (Y.H.); qxtdmj@aliyun.com (X.Q.); cyydmj@tom.com (Y.C.); qsmdmj@tom.com (S.Q.) 2 Qingdao Bolong Experimental Animal Co., Ltd., Qingdao 266225, China; wdddmj@126.com (D.W.); sjdmja@126.com (J.S.) 3 China Ostrich Farming and Development Association, Beijing 100026, China; diyouxiayouxiang@163.com (Y.D.); wangxiaoyiyx2020@163.com (X.W.); ssgyouxiang2020@163.com (S.S.); wph2020@163.com (P.W.); xslyx2020@163.com (S.X.) 4 College of Biological Sciences, China Agricultural University, Beijing 100193, China; yylyx2020@163.com * Correspondence: weiquan8@126.com (W.L.); yanboyin2011@163.com (Y.Y.) Received: 15 January 2020; Accepted: 12 March 2020; Published: 19 March 2020 Abstract: Ostrich diseases characterized by paralysis have been breaking out in broad areas of China since 2015, causing major damage to the ostrich breeding industry in China. This report describes a parvovirus detected in ostriches from four difffferent regions. The entire genomes of four parvovirus strains were sequenced following amplifification by PCR, and we conducted comprehensive analysis of the ostrich parvovirus genome. Results showed that the length genomes of the parvovirus contained two open reading frames. Ostrich parvovirus (OsPV) is a branch of goose parvovirus (GPV). Genetic distance analysis revealed a close relationship between the parvovirus and goose parvovirus strains from China, with the closest being the 2016 goose parvovirus RC16 strain from Chongqing. This is the fifirst report of a parvovirus in ostriches. However, whether OsPV is the pathogen of ostrich paralysis remains uncertain. This study contributes new information about the evolution and epidemiology of parvovirus in China, which provides a new way for the study of paralysis in ostriches. Keywords: ostrich; leg paralysis; parvovirus; phylogenetic analysis; whole genome amplifification 1. Introduction Parvoviruses are small, non-enveloped, linear single-stranded DNA viruses [1]. Parvoviruses may have appeared millions of years ago, infecting invertebrates and vertebrates [2]. All parvoviruses that infect vertebrates belong to the parvovirinae subfamily [3]. Parvoviruses are widespread in birds; parvoviruses that cause harm to the health of avians mainly include goose parvovirus (GPV), Muscovy duck parvovirus (MDPV), and chicken and turkey parvoviruses [2]. Recent studies have shown that the diversity of known parvovirus species has greatly expanded and the host range of parvovirus may include the entire animal kingdom [4]. In recent years, an outbreak of disease has occurred in farmed ostriches aged from 1 to 4 months, with paralysis as the main clinical manifestation, and an incidence rate ranging from 30% to 80%. Sick ostriches gradually become thin and weak, with the disease lasting for about one month from onset to death. However, no visceral lesions have been found by gross examination in the heart, liver, lung, Viruses 2020, 12, 334; doi:10.3390/v12030334 www.mdpi.com/journal/virusesViruses 2020, 12, 334 2 of 9 kidney, or spleen of sick ostriches. Small bleeding points in leg muscles, accompanied by increased joint flfluids, have been observed in a few animals, but no visible lesions have been detected in most infected ostriches. Antibacterial drugs and antiviral drugs, such as ribavirin, have been ineffffective in the treatment of the disease. While paralysis of nestlings has caused extensive loss within the ostrich farming industry in China, no effffective solution has been reported so far. Here, we describe a hitherto unknown ostrich parvovirus (OsPV), detected in paralyzed ostrich nestlings from the Beijing municipality and Hebei, Shanxi, and Yunnan provinces. Whole genomic sequencing and phylogenetic analyses have been carried out on strains, from each region, and their molecular evolutionary relationship with goose parvovirus (GPV) and Muscovy duck parvovirus (MDPV) established. 2. Materials and Methods 2.1. Sample Collection and PCR Detection Sources of pathological materials were paralyzed ostrich nestlings from difffferent farms in Beijing, Yunnan, Shanxi, and Hebei (Table 1). Livers, spleens, hearts, and brain tissues were collected from dead animals, and samples were homogenized in PBS, followed by freezing and thawing (3 times) and centrifugation for the collection of supernatants. DNA was extracted using a MiniBEST Viral RNA/DNA Extraction Kit Ver.5.0 (Takara, Beijing, China), according to the manufacturer’s instructions. The DNA of the samples was preserved at t 20 ◦C. PCR was carried out according to the procedure of Tatár-Kis et al. [5], and the primers were according to the method of Zadori Z et al. [6]. All of the positive products were sequenced by Ruibiotech, Qingdao, China. Sampling was carried out by a veterinarian, who took difffferent samples as part of his routine work and under the permission of the farm owner. For this reason, sampling did not require the approval of the Ethics Committee.
Table 1. Detection results of ostrich parvovirus (OsPV) in clinical samples.
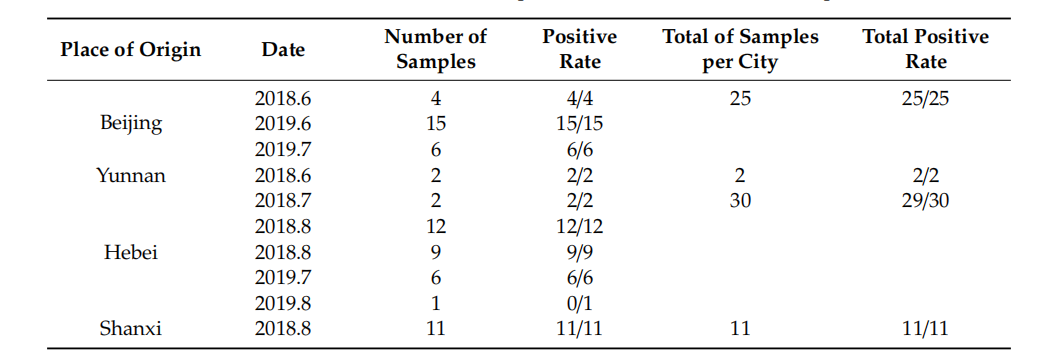
2.2. Whole Genome Amplifification of OsPV The primers were designed as described by Li et al. [7] to amplify the complete genomes of the OsPV strains. The genomes were amplifified using PrimeStar HS DNA polymerase (TaKaRa, Beijng, China). PCR conditions were as follows: initial denaturation at 95 ◦C for 5 min, followed by 33 cycles of 95 ◦C for 45 s, 55 ◦C for 45 s, 72 ◦C for 60 s, and termination for 10 min at 72 ◦C. Amplifified DNA fragments were visualized after electrophoresis on a 1% agarose gel (Tsingke, Beijng, China). PCR products were purifified using a PCR purifification kit (Cwbiotech, Beijng, China)., cloned into the pMD18-T vector (Takara, Beijing, China), and were sequenced by Ruibiotech, Qingdao.Viruses 2020, 12, 334 3 of 9 2.3. Sequence Alignment and Phylogenetic Analysis Sequence alignment and homology comparison based on nucleotide sequences between the four OsPVs obtained in this study, and other parvovirus sequences published in GenBank (Table S1), were conducted using Clustal W within the MegAlign program (DNASTAR Inc., Madison, WI, USA). and the MEGA program was used for pairwise distance analysis Proteins with motifs and domains were sought using profifile hidden Markov models deployed by the HMMER server (https: //www.ebi.ac.uk/Tools/hmmer/). RDP version 4.0 software was used for analyzing the recombination in the four OsPVs, to understand the relationship of OsPVs with GPV, MDPV. Phylogenetic trees based on replication (Rep) protein and capsid (Cap) protein sequences was constructed by using the maximum likelihood method with a Poisson model, based on 1000 bootstrap duplicates. Bootstrap values > 70% were considered to be signifificant. The nucleotides of OsPVs and classical avian parvovirus were compared by MegAlign, and the homology between OsPV and other avian parvovirus was analyzed. 3. Results Parvoviral DNA was detected in a number of tissue samples (Table 1). Four positive samples, from Yunnan (OsPV-YN, Genbank Accession No.MK281604), Shanxi (OsPV-YQ, Genbank Accession No. MK281605), Hebei (OsPV-SJZ, Genbank Accession No. MK281603), and Beijing (OsPV-BJ, Genbank Accession No. MK281602) were sequenced. Results showed that the OsPV genome contained two major open reading frames (ORFs), which were similar to goose and Muscovy duck parvoviruses. The genome contains two open reading frames (ORFs), which can be divided into left ORF and right ORF. The left ORF encodes two replication (Rep) proteins, Rep1 and Rep2 [7]. The right ORF encodes three capsid (Cap) proteins named VP1, VP2, and VP3 [6]. The length genomes ranged from 5041–5103 nt containing 416–446 nt inverted terminal repeats (ITRs). The left ORF contained 1884 nt that encoding the Rep protein with 628 amino acids, while the right ORF of 2199 nt coded for the Cap protein with 733 amino acids. The nucleotide homology of the four OsPV genomes ranged from 99.0–99.4%. The nucleotide homology of the encoded Cap protein was 98.8–99.4% while that of the encoded Rep protein was 99.0–99.7%. The highest homology, 99.4%, was between the Shanxi and Hebei strains. 3.1. Gene Homology Comparisons with Other Avian Parvoviruses Table 2 provides a gene homology comparison of OsPV, GPV, NGPV (parvovirus isolated from duck tongue disease), and MDPV. The OsPVs showed higher homology with GPV and NGPV—especially the GPV RC16 strain—than with MDPV.
Table 2. Gene homology comparisons with other avian parvoviruses.
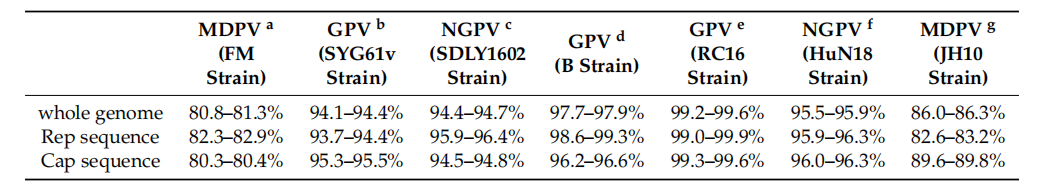
a. The MDPV FM strain of Muscovy duck parvovirus (MDPV), discovered in Hungary in 1993 by Zadori et al. [6]; b. SYG61v strain (vaccine strain) of goose parvovirus (GPV); c. novel goose parvovirus-related virus (NGPV) strain SDLY1602 responsible for duck tongue disease, found in Shandong province in 2016 by Li et al. [7]; d. GPV strain B isolated from grey goose in Hungary [6]; e. GPV-RC16 strain from Chongqing reported in 2017 by Liu et al. [1,8]; f. the NGPV HuN18 strain of Novel goose parvovirus (NGPV), discovered in Hunan province in 2018 by Wan C et al. [9]; g. MDPV strain JH10 isolated from Muscovy duck in China [10].